Caution
You're reading the documentation for a development version. For the latest released version, please have a look at 0.11.0.
librosa.decompose.decompose
- librosa.decompose.decompose(S, *, n_components=None, transformer=None, sort=False, fit=True, **kwargs)[source]
Decompose a feature matrix.
Given a spectrogram
S, produce a decomposition intocomponentsandactivationssuch thatS ~= components.dot(activations).By default, this is done with with non-negative matrix factorization (NMF), but any
sklearn.decomposition-type object will work.- Parameters:
- Snp.ndarray [shape=(…, n_features, n_samples), dtype=float]
The input feature matrix (e.g., magnitude spectrogram)
If the input has multiple channels (leading dimensions), they will be automatically flattened prior to decomposition.
If the input is multi-channel, channels and features are automatically flattened into a single axis before the decomposition. For example, a stereo input S with shape (2, n_features, n_samples) is automatically reshaped to (2 * n_features, n_samples).
- n_componentsint > 0 [scalar] or None
number of desired components
if None, then
n_featurescomponents are used- transformerNone or object
If None, use
sklearn.decomposition.NMFOtherwise, any object with a similar interface to NMF should work.
transformermust follow the scikit-learn convention, where input data is(n_samples, n_features).transformer.fit_transform() will be run on
S.T(notS), the return value of which is stored (transposed) asactivationsThe components will be retrieved as
transformer.components_.T:S ~= np.dot(activations, transformer.components_).T
or equivalently:
S ~= np.dot(transformer.components_.T, activations.T)
- sortbool
If
True, components are sorted by ascending peak frequency.Note
If used with
transformer, sorting is applied to copies of the decomposition parameters, and not totransformerinternal parameters.Warning
If the input array has more than two dimensions (e.g., if it’s a multi-channel spectrogram), then axis sorting is not supported and a
ParameterErrorexception is raised.- fitbool
If True, components are estimated from the input
S.If False, components are assumed to be pre-computed and stored in
transformer, and are not changed.- **kwargsAdditional keyword arguments to the default transformer
- Returns:
- components: np.ndarray [shape=(…, n_features, n_components)]
matrix of components (basis elements).
- activations: np.ndarray [shape=(n_components, n_samples)]
transformed matrix/activation matrix
- Raises:
- ParameterError
if
fitis False and notransformerobject is provided.if the input array is multi-channel and
sort=Trueis specified.
See also
sklearn.decompositionSciKit-Learn matrix decomposition modules
Examples
Decompose a magnitude spectrogram into 16 components with NMF
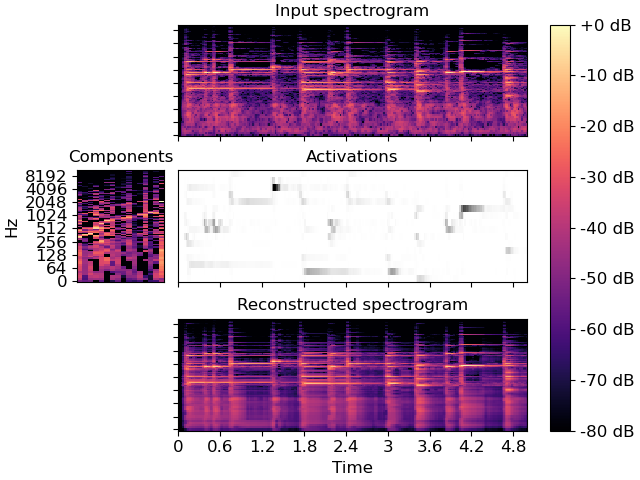
>>> y, sr = librosa.load(librosa.ex('pistachio'), duration=5) >>> S = np.abs(librosa.stft(y)) >>> comps, acts = librosa.decompose.decompose(S, n_components=16)
Sort components by ascending peak frequency
>>> comps, acts = librosa.decompose.decompose(S, n_components=16, ... sort=True)
Or with sparse dictionary learning
>>> import sklearn.decomposition >>> T = sklearn.decomposition.MiniBatchDictionaryLearning(n_components=16) >>> scomps, sacts = librosa.decompose.decompose(S, transformer=T, sort=True)
>>> import matplotlib.pyplot as plt >>> layout = [list(".AAAA"), list("BCCCC"), list(".DDDD")] >>> fig, ax = plt.subplot_mosaic(layout, constrained_layout=True) >>> librosa.display.specshow(librosa.amplitude_to_db(S, ref=np.max), ... y_axis='log', x_axis='time', ax=ax['A']) >>> ax['A'].set(title='Input spectrogram') >>> ax['A'].label_outer() >>> librosa.display.specshow(librosa.amplitude_to_db(comps, >>> ref=np.max), >>> y_axis='log', ax=ax['B']) >>> ax['B'].set(title='Components') >>> ax['B'].label_outer() >>> ax['B'].sharey(ax['A']) >>> librosa.display.specshow(acts, x_axis='time', ax=ax['C'], cmap='gray_r') >>> ax['C'].set(ylabel='Components', title='Activations') >>> ax['C'].sharex(ax['A']) >>> ax['C'].label_outer() >>> S_approx = comps.dot(acts) >>> img = librosa.display.specshow(librosa.amplitude_to_db(S_approx, >>> ref=np.max), >>> y_axis='log', x_axis='time', ax=ax['D']) >>> ax['D'].set(title='Reconstructed spectrogram') >>> ax['D'].sharex(ax['A']) >>> ax['D'].sharey(ax['A']) >>> ax['D'].label_outer() >>> fig.colorbar(img, ax=list(ax.values()), format="%+2.f dB")