Caution
You're reading an old version of this documentation. If you want up-to-date information, please have a look at 0.9.1.
Note
Click here to download the full example code
Laplacian segmentation¶
This notebook implements the laplacian segmentation method of McFee and Ellis, 2014, with a couple of minor stability improvements.
Throughout the example, we will refer to equations in the paper by number, so it will be helpful to read along.
# Code source: Brian McFee
# License: ISC
- Imports
numpy for basic functionality
scipy for graph Laplacian
matplotlib for visualization
sklearn.cluster for K-Means
from __future__ import print_function
import numpy as np
import scipy
import matplotlib.pyplot as plt
import sklearn.cluster
import librosa
import librosa.display
First, we’ll load in a song
Out:
/tmp/tmpfl4ra6qp/b0064fe7dbe8048b1d4148e61a568b6fe3fca91b/librosa/core/audio.py:161: UserWarning: PySoundFile failed. Trying audioread instead.
warnings.warn('PySoundFile failed. Trying audioread instead.')
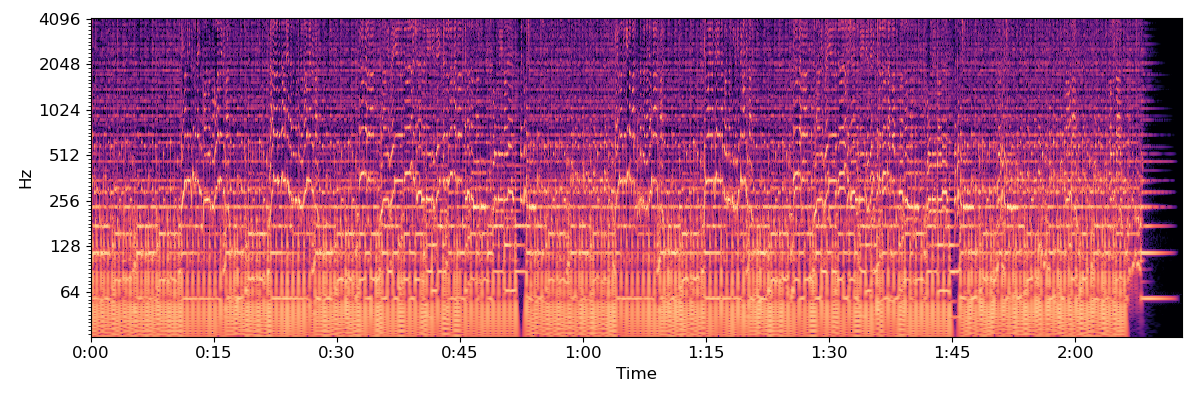
Next, we’ll compute and plot a log-power CQT
BINS_PER_OCTAVE = 12 * 3
N_OCTAVES = 7
C = librosa.amplitude_to_db(np.abs(librosa.cqt(y=y, sr=sr,
bins_per_octave=BINS_PER_OCTAVE,
n_bins=N_OCTAVES * BINS_PER_OCTAVE)),
ref=np.max)
plt.figure(figsize=(12, 4))
librosa.display.specshow(C, y_axis='cqt_hz', sr=sr,
bins_per_octave=BINS_PER_OCTAVE,
x_axis='time')
plt.tight_layout()
Out:
/tmp/tmpfl4ra6qp/b0064fe7dbe8048b1d4148e61a568b6fe3fca91b/librosa/display.py:862: MatplotlibDeprecationWarning: The 'basey' parameter of __init__() has been renamed 'base' since Matplotlib 3.3; support for the old name will be dropped two minor releases later.
scaler(mode, **kwargs)
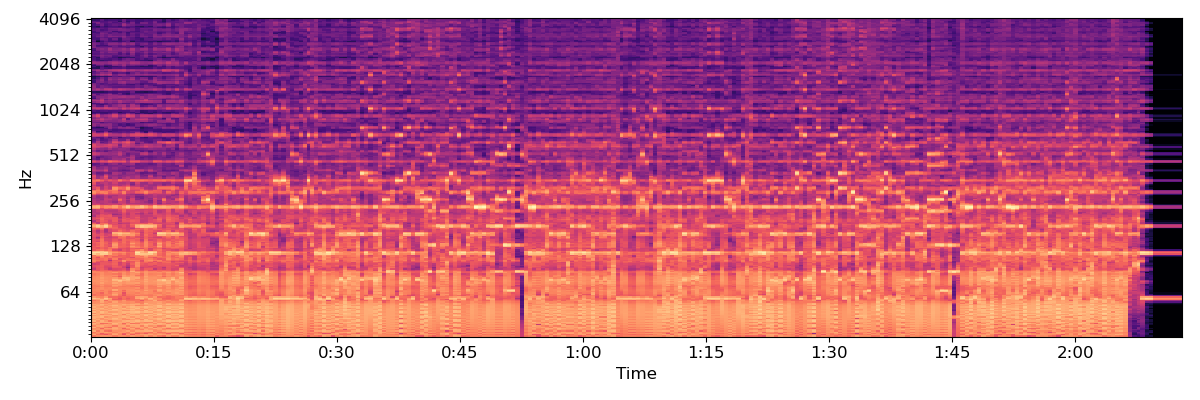
To reduce dimensionality, we’ll beat-synchronous the CQT
tempo, beats = librosa.beat.beat_track(y=y, sr=sr, trim=False)
Csync = librosa.util.sync(C, beats, aggregate=np.median)
# For plotting purposes, we'll need the timing of the beats
# we fix_frames to include non-beat frames 0 and C.shape[1] (final frame)
beat_times = librosa.frames_to_time(librosa.util.fix_frames(beats,
x_min=0,
x_max=C.shape[1]),
sr=sr)
plt.figure(figsize=(12, 4))
librosa.display.specshow(Csync, bins_per_octave=12*3,
y_axis='cqt_hz', x_axis='time',
x_coords=beat_times)
plt.tight_layout()
Out:
/tmp/tmpfl4ra6qp/b0064fe7dbe8048b1d4148e61a568b6fe3fca91b/librosa/display.py:862: MatplotlibDeprecationWarning: The 'basey' parameter of __init__() has been renamed 'base' since Matplotlib 3.3; support for the old name will be dropped two minor releases later.
scaler(mode, **kwargs)
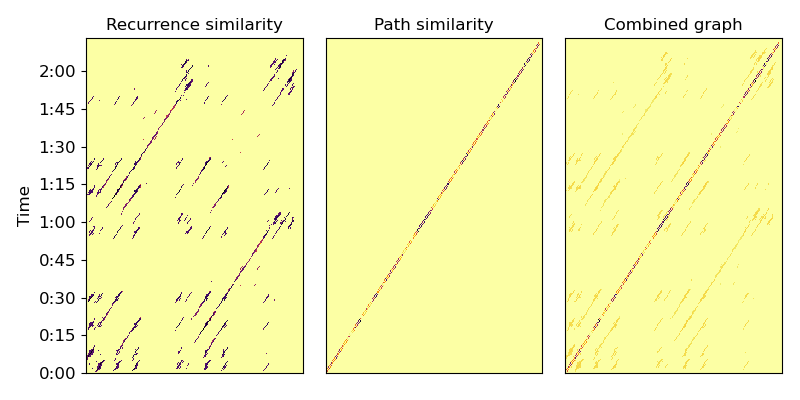
Let’s build a weighted recurrence matrix using beat-synchronous CQT (Equation 1) width=3 prevents links within the same bar mode=’affinity’ here implements S_rep (after Eq. 8)
R = librosa.segment.recurrence_matrix(Csync, width=3, mode='affinity',
sym=True)
# Enhance diagonals with a median filter (Equation 2)
df = librosa.segment.timelag_filter(scipy.ndimage.median_filter)
Rf = df(R, size=(1, 7))
Now let’s build the sequence matrix (S_loc) using mfcc-similarity
\(R_\text{path}[i, i\pm 1] = \exp(-\|C_i - C_{i\pm 1}\|^2 / \sigma^2)\)
Here, we take \(\sigma\) to be the median distance between successive beats.
And compute the balanced combination (Equations 6, 7, 9)
Plot the resulting graphs (Figure 1, left and center)
plt.figure(figsize=(8, 4))
plt.subplot(1, 3, 1)
librosa.display.specshow(Rf, cmap='inferno_r', y_axis='time',
y_coords=beat_times)
plt.title('Recurrence similarity')
plt.subplot(1, 3, 2)
librosa.display.specshow(R_path, cmap='inferno_r')
plt.title('Path similarity')
plt.subplot(1, 3, 3)
librosa.display.specshow(A, cmap='inferno_r')
plt.title('Combined graph')
plt.tight_layout()
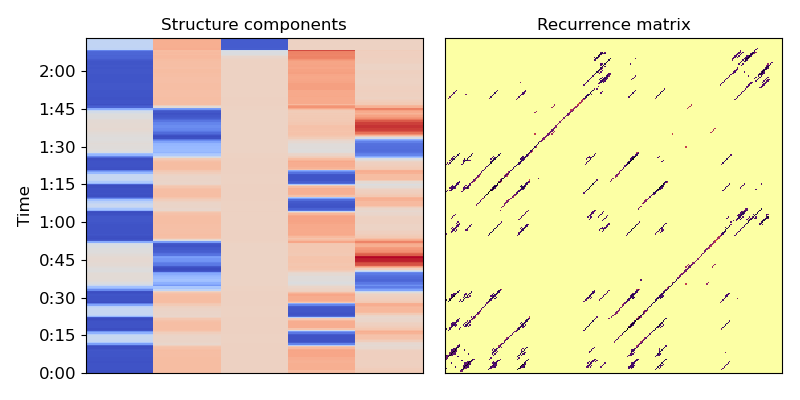
Now let’s compute the normalized Laplacian (Eq. 10)
L = scipy.sparse.csgraph.laplacian(A, normed=True)
# and its spectral decomposition
evals, evecs = scipy.linalg.eigh(L)
# We can clean this up further with a median filter.
# This can help smooth over small discontinuities
evecs = scipy.ndimage.median_filter(evecs, size=(9, 1))
# cumulative normalization is needed for symmetric normalize laplacian eigenvectors
Cnorm = np.cumsum(evecs**2, axis=1)**0.5
# If we want k clusters, use the first k normalized eigenvectors.
# Fun exercise: see how the segmentation changes as you vary k
k = 5
X = evecs[:, :k] / Cnorm[:, k-1:k]
# Plot the resulting representation (Figure 1, center and right)
plt.figure(figsize=(8, 4))
plt.subplot(1, 2, 2)
librosa.display.specshow(Rf, cmap='inferno_r')
plt.title('Recurrence matrix')
plt.subplot(1, 2, 1)
librosa.display.specshow(X,
y_axis='time',
y_coords=beat_times)
plt.title('Structure components')
plt.tight_layout()
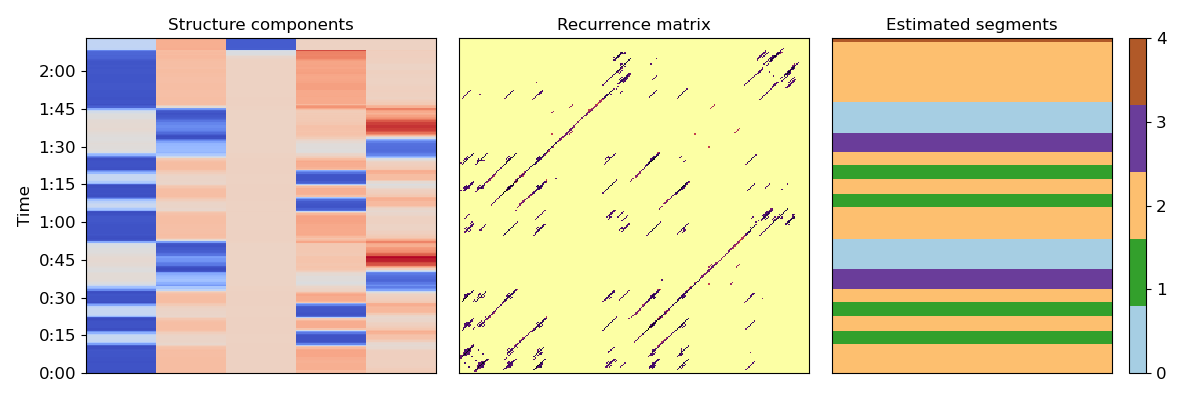
Let’s use these k components to cluster beats into segments (Algorithm 1)
KM = sklearn.cluster.KMeans(n_clusters=k)
seg_ids = KM.fit_predict(X)
# and plot the results
plt.figure(figsize=(12, 4))
colors = plt.get_cmap('Paired', k)
plt.subplot(1, 3, 2)
librosa.display.specshow(Rf, cmap='inferno_r')
plt.title('Recurrence matrix')
plt.subplot(1, 3, 1)
librosa.display.specshow(X,
y_axis='time',
y_coords=beat_times)
plt.title('Structure components')
plt.subplot(1, 3, 3)
librosa.display.specshow(np.atleast_2d(seg_ids).T, cmap=colors)
plt.title('Estimated segments')
plt.colorbar(ticks=range(k))
plt.tight_layout()
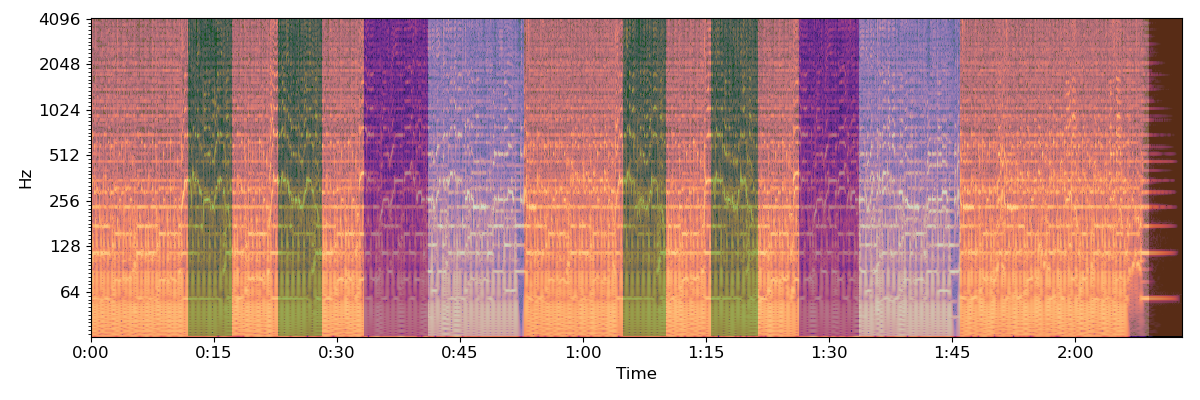
Locate segment boundaries from the label sequence
bound_beats = 1 + np.flatnonzero(seg_ids[:-1] != seg_ids[1:])
# Count beat 0 as a boundary
bound_beats = librosa.util.fix_frames(bound_beats, x_min=0)
# Compute the segment label for each boundary
bound_segs = list(seg_ids[bound_beats])
# Convert beat indices to frames
bound_frames = beats[bound_beats]
# Make sure we cover to the end of the track
bound_frames = librosa.util.fix_frames(bound_frames,
x_min=None,
x_max=C.shape[1]-1)
And plot the final segmentation over original CQT
# sphinx_gallery_thumbnail_number = 5
import matplotlib.patches as patches
plt.figure(figsize=(12, 4))
bound_times = librosa.frames_to_time(bound_frames)
freqs = librosa.cqt_frequencies(n_bins=C.shape[0],
fmin=librosa.note_to_hz('C1'),
bins_per_octave=BINS_PER_OCTAVE)
librosa.display.specshow(C, y_axis='cqt_hz', sr=sr,
bins_per_octave=BINS_PER_OCTAVE,
x_axis='time')
ax = plt.gca()
for interval, label in zip(zip(bound_times, bound_times[1:]), bound_segs):
ax.add_patch(patches.Rectangle((interval[0], freqs[0]),
interval[1] - interval[0],
freqs[-1],
facecolor=colors(label),
alpha=0.50))
plt.tight_layout()
plt.show()
Out:
/tmp/tmpfl4ra6qp/b0064fe7dbe8048b1d4148e61a568b6fe3fca91b/librosa/display.py:862: MatplotlibDeprecationWarning: The 'basey' parameter of __init__() has been renamed 'base' since Matplotlib 3.3; support for the old name will be dropped two minor releases later.
scaler(mode, **kwargs)
Total running time of the script: ( 0 minutes 12.621 seconds)